Identify presence/absence variants
Author: Andrea Guarracino
Synopsis
The term presence/absence variation (PAV) is used to describe sequences that are present in one genome, but
entirely missing in another genome, and is an important source of genetic divergence and diversity. odgi pav allows
users to identify PAVs with respect to one or multiple paths in the pangenome graph.
Steps
Build the Lipoprotein A graph
Assuming that your current working directory is the root of the odgi project, to construct an odgi file from the
LPA dataset in GFA format, execute:
odgi build -g test/LPA.gfa -o LPA.og
The command creates a file called LPA.og, which contains the input graph in odgi format. This graph contains
13 contigs from 7 haploid human genome assemblies from 6 individuals plus the chm13 cell line. The contigs cover the
Lipoprotein A (LPA) locus, which encodes the
Apo(a) protein.
Presence/absence variants (PAVs)
Any path in the graph can be used as a reference to identify PAVs. In this example, we have chosen the chm13__LPA__tig00000001
path. To obtain 1000 bp interval windows across the chosen reference, execute:
odgi paths -i LPA.og -f | grep 'chm13__LPA__tig00000001' -A 1 > reference.fa
samtools faidx reference.fa
bedtools makewindows -g <(cut -f 1,2 reference.fa.fai) -w 1000 > LPA.w1kbp.bed
To identify the PAVs, execute:
odgi pav -i LPA.og -b LPA.w1kbp.bed > LPA.w1kbp.pavs.tsv
By default, odgi pav prints to stdout a TSV table with the PAV ratios.
For a given path range PR and path P, the PAV ratio is the ratio between the sum of the lengths of the nodes
in PR that are crossed by P divided by the sum of the lengths of all the nodes in PR.
Each node is considered only once.
To take a look at the first rows of the table in the LPA.w1kbp.pavs.tsv file, execute:
head LPA.w1kbp.pavs.tsv | column -t
chrom start end name group pav
chm13__LPA__tig00000001 0 1000 . chm13__LPA__tig00000001 1
chm13__LPA__tig00000001 0 1000 . HG002__LPA__tig00000001 0
chm13__LPA__tig00000001 0 1000 . HG002__LPA__tig00000005 0
chm13__LPA__tig00000001 0 1000 . HG00733__LPA__tig00000001 0
chm13__LPA__tig00000001 0 1000 . HG00733__LPA__tig00000008 0
chm13__LPA__tig00000001 0 1000 . HG01358__LPA__tig00000002 0
chm13__LPA__tig00000001 0 1000 . HG01358__LPA__tig00000010 0
chm13__LPA__tig00000001 0 1000 . HG02572__LPA__tig00000005 0.99524
chm13__LPA__tig00000001 0 1000 . HG02572__LPA__tig00000001 0.99524
The chrom, start, end, and name columns are filled with the values in the corresponding columns in the
input BED format file. In this example, the region chm13__LPA__tig00000001:0-1000 is covered at 99.54% in the
HG02572__LPA__tig00000005 and HG02572__LPA__tig00000001 contigs, and it is absent in the others, except in the
reference itself.
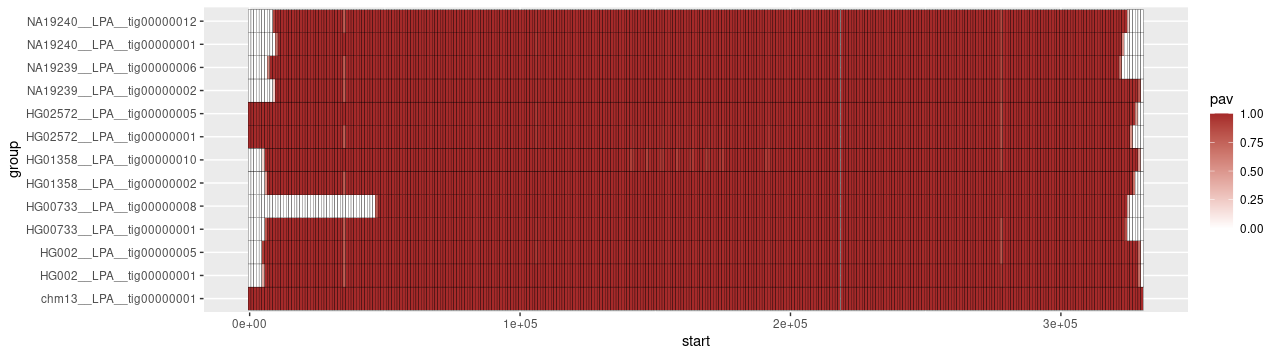
To display the result, execute
library(tidyverse)
pav_table <- read.table('LPA.w1kbp.pavs.tsv', sep = '\t', header = T)
pav_table %>%
ggplot(aes(x = start, y = group, fill = pav)) +
geom_tile(color = "black") +
scale_fill_gradient(low = "white", high = "brown")
to obtain the following visualization:
odgi pav also supports the matrix format output (-M/matrix-output flag).
To emit the PAV ratios in a matrix and take a look at its first rows and columns, execute:
odgi pav -i LPA.og -b LPA.w1kbp.bed -M > LPA.w1kbp.pavs.matrix.txt
head LPA.w1kbp.pavs.matrix.txt | cut -f 1-8 | column -t
chrom start end name chm13__LPA__tig00000001 HG002__LPA__tig00000001 HG002__LPA__tig00000005 HG00733__LPA__tig00000001
chm13__LPA__tig00000001 0 1000 . 1 0 0 0
chm13__LPA__tig00000001 1000 2000 . 1 0 0 0
chm13__LPA__tig00000001 2000 3000 . 1 0 0 0
chm13__LPA__tig00000001 3000 4000 . 1 0 0 0
chm13__LPA__tig00000001 4000 5000 . 1 0 0 0
chm13__LPA__tig00000001 5000 6000 . 1 0.4156 0.91101 0.00091743
chm13__LPA__tig00000001 6000 7000 . 1 1 1 0.80339
chm13__LPA__tig00000001 7000 8000 . 1 0.99811 0.99906 0.98491
chm13__LPA__tig00000001 8000 9000 . 1 1 1 0.99466
To emit a binary PAV matrix, execute:
odgi pav -i LPA.og -b LPA.w1kbp.bed -M -B 0.5 > LPA.w1kbp.pavs.matrix.binary.txt
head LPA.w1kbp.pavs.matrix.binary.txt | cut -f 1-8 | column -t
chrom start end name chm13__LPA__tig00000001 HG002__LPA__tig00000001 HG002__LPA__tig00000005 HG00733__LPA__tig00000001
chm13__LPA__tig00000001 0 1000 . 1 0 0 0
chm13__LPA__tig00000001 1000 2000 . 1 0 0 0
chm13__LPA__tig00000001 2000 3000 . 1 0 0 0
chm13__LPA__tig00000001 3000 4000 . 1 0 0 0
chm13__LPA__tig00000001 4000 5000 . 1 0 0 0
chm13__LPA__tig00000001 5000 6000 . 1 0 1 0
chm13__LPA__tig00000001 6000 7000 . 1 1 1 1
chm13__LPA__tig00000001 7000 8000 . 1 1 1 1
chm13__LPA__tig00000001 8000 9000 . 1 1 1 1
With B is specified to emit a binary matrix, with 1 if the PAV ratio is greater than or equal to the specified
threshold (0.5 in the example), else 0.
If needed, it is possible to group paths. For this, we need to prepare a file that specifies for each path the group it
belongs to. In the LPA pangenome graph, the first part of each path name indicates the sample name. Therefore, to
prepare such a file, execute:
odgi paths -i LPA.og -L > LPA.paths.txt
cut -f 1 -d '_' LPA.paths.txt > LPA.samples.txt
paste LPA.paths.txt LPA.samples.txt > LPA.path_and_sample.txt
head LPA.path_and_sample.txt -n 5 | column -t
chm13__LPA__tig00000001 chm13
HG002__LPA__tig00000001 HG002
HG002__LPA__tig00000005 HG002
HG00733__LPA__tig00000001 HG00733
HG00733__LPA__tig00000008 HG00733
Then, to group the PAVs by sample, execute:
odgi pav -i LPA.og -b LPA.w1kbp.bed -M -B 0.5 -p LPA.path_and_sample.txt > LPA.w1kbp.pavs.matrix.binary.grouped_by_sample.txt
head LPA.w1kbp.pavs.matrix.binary.grouped_by_sample.txt | column -t
chrom start end name HG002 HG00733 HG01358 HG02572 NA19239 NA19240 chm13
chm13__LPA__tig00000001 0 1000 . 0 0 0 1 0 0 1
chm13__LPA__tig00000001 1000 2000 . 0 0 0 1 0 0 1
chm13__LPA__tig00000001 2000 3000 . 0 0 0 1 0 0 1
chm13__LPA__tig00000001 3000 4000 . 0 0 0 1 0 0 1
chm13__LPA__tig00000001 4000 5000 . 0 0 0 1 0 0 1
chm13__LPA__tig00000001 5000 6000 . 1 0 0 1 0 0 1
chm13__LPA__tig00000001 6000 7000 . 1 1 1 1 0 0 1
chm13__LPA__tig00000001 7000 8000 . 1 1 1 1 1 0 1
chm13__LPA__tig00000001 8000 9000 . 1 1 1 1 1 0 1
How to get a BED file: odgi untangle
Instead of splitting in windows the path(s) chosen as a reference(s), an alternative way to obtain a BED file for odgi pav
is to use odgi untangle (see the corresponding tutorial Untangling the pangenome for more information on how it works).
For example, to identify the PAVs by considering chm13__LPA__tig00000001 as reference path, execute:
odgi untangle -i LPA.og -r chm13__LPA__tig00000001 | sed '1d' | cut -f 4,5,6 | sort | uniq | sort -k 2n > LPA.untangle.bed
odgi pav -i LPA.og -b LPA.untangle.bed > LPA.untangle.pavs.tsv
head LPA.untangle.pavs.tsv | head -n 5 | column -t
chrom start end name group pav
chm13__LPA__tig00000001 0 5045 . chm13__LPA__tig00000001 1
chm13__LPA__tig00000001 0 5045 . HG002__LPA__tig00000001 0
chm13__LPA__tig00000001 0 5045 . HG002__LPA__tig00000005 0
chm13__LPA__tig00000001 0 5045 . HG00733__LPA__tig00000001 0
Of note, odgi pav is not constrained to use a single reference. As further example, to identify the PAVs by considering
all paths as reference paths and emit them in a matrix, execute:
odgi paths -i LPA.og -L > LPA.paths.txt
odgi untangle -i LPA.og -R LPA.paths.txt | sed '1d' | cut -f 4,5,6 | sort | uniq > LPA.untangle.multiple_references.bed
odgi pav -i LPA.og -b LPA.untangle.multiple_references.bed -M > LPA.untangle.multiple_references.pavs.matrix.txt
# Sort by starting position, but keeping the header line at the top
awk 'NR == 1; NR > 1 {print $0 | "sort -k 2n"}' LPA.untangle.multiple_references.pavs.matrix.txt | head | cut -f 1-8 | column -t
chrom start end name chm13__LPA__tig00000001 HG002__LPA__tig00000001 HG002__LPA__tig00000005 HG00733__LPA__tig00000001
chm13__LPA__tig00000001 0 5045 . 1 0 0 0
HG002__LPA__tig00000001 0 241 . 0.99585 1 0.9917 0
HG002__LPA__tig00000005 0 540 . 1 0 1 0
HG00733__LPA__tig00000001 0 403 . 0.98263 0.98263 0.98263 1
HG00733__LPA__tig00000008 0 93388 . 0.99935 0.99954 0.99769 0.99908
HG01358__LPA__tig00000002 0 880 . 0.99886 0.99773 0.99886 0.98068
HG02572__LPA__tig00000001 0 35 . 0 0 0 0
NA19239__LPA__tig00000006 0 1665 . 1 0.9994 0.9994 0.99219
NA19240__LPA__tig00000001 0 36676 . 0.99954 0.98871 0.98901 0.98849
How to get a BED file: odgi flatten
Similarly, we can obtain a BED file for odgi pav also by applying odgi flatten. For example, to identify the PAVs
for all nodes crossed by all paths in the graph, execute:
odgi flatten -i LPA.og -b LPA.flatten.tsv
sed '1d' LPA.flatten.tsv | awk -v OFS='\t' '{print($4,$2,$3,"step.rank_"$6,".",$5)}' > LPA.flatten.bed
odgi pav -i LPA.og -b LPA.flatten.bed > LPA.flatten.pavs.tsv
head LPA.flatten.pavs.tsv | column -t
chrom start end name group pav
chm13__LPA__tig00000001 38 43 step.rank_0 chm13__LPA__tig00000001 1
chm13__LPA__tig00000001 38 43 step.rank_0 HG002__LPA__tig00000001 0
chm13__LPA__tig00000001 38 43 step.rank_0 HG002__LPA__tig00000005 0
chm13__LPA__tig00000001 38 43 step.rank_0 HG00733__LPA__tig00000001 0